Note
Go to the end to download the full example code.
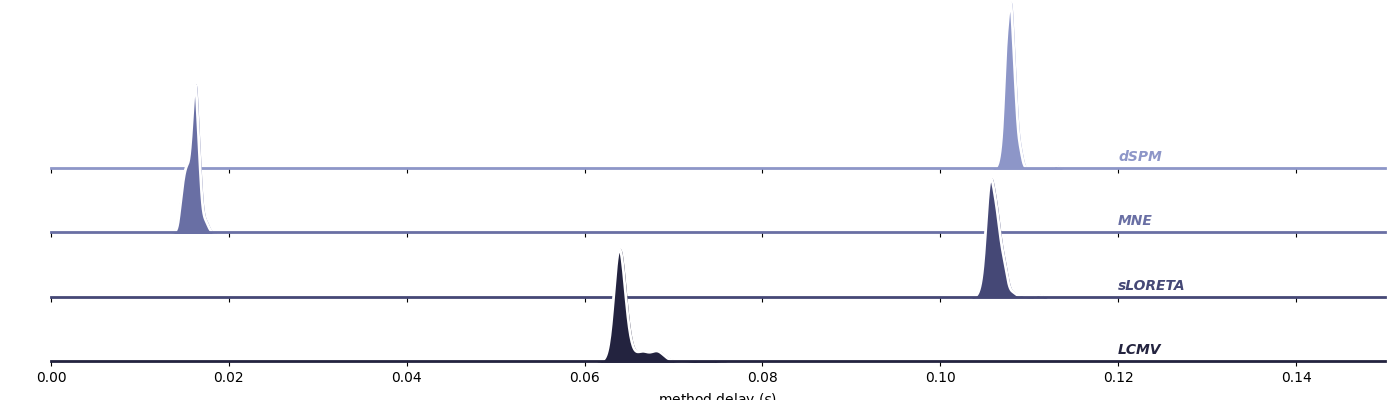
Compare Delays Across Different Source Localization Methods#
This example visually compares the delays observed for several common source localization techniques:
MNE - Minimum Norm Estimate
dSPM - Dynamic Statistical Parametric Mapping
eLORETA - Exact Low-Resolution Electromagnetic Tomography
sLORETA - Standardized Low-Resolution Electromagnetic Tomography
LCMV - Linearly Constrained Minimum Variance beamformer
Now let’s load data and visualize each method’s delay.
Helper function for labeling each row
def label(x, color, label):
ax = plt.gca()
ax.text(0.8, .2, label, fontweight="bold", fontstyle='italic', color=color,
ha="left", va="center", transform=ax.transAxes)
Function to plot KDEs for a list of methods
def plot_method_delays(df, method_names, xlim, ylim, bw_adjust=1, top=0.72):
"""
Plot KDEs of delays for specified neurofeedback methods.
Parameters
----------
df : pandas.DataFrame
Must contain columns 'method' and 'delay'.
method_names : list of str
Methods to plot.
xlim : list of float
X-axis limits.
ylim : list of float
Y-axis limits.
bw_adjust : float, optional
Bandwidth adjustment for KDE (default 1).
top : float, optional
Top position for FacetGrid (default 0.72).
Returns
-------
g : seaborn.FacetGrid
FacetGrid object.
"""
df_sub = df.query("method == @method_names")
pal = sns.cubehelix_palette(len(method_names), rot=-.05, light=.6)
g = sns.FacetGrid(
df_sub, row="method", hue="method", aspect=14, height=1,
palette=pal, row_order=method_names, xlim=xlim, ylim=ylim
)
g.map(sns.kdeplot, "delay", bw_adjust=bw_adjust, clip_on=False, clip=xlim,
fill=True, alpha=1, linewidth=1.5)
g.map(sns.kdeplot, "delay", clip_on=False, color="w", clip=xlim,
lw=2, bw_adjust=bw_adjust)
g.refline(y=0, linewidth=2, linestyle="-", color=None, clip_on=False)
g.map(label, "delay")
g.figure.subplots_adjust(hspace=.15, top=top)
g.set_titles("")
g.set(yticks=[], ylabel="", xlabel=r"method delay ($s$)")
g.despine(bottom=True, left=True)
return g
Next, visualize delays for “dSPM”, “MNE”, “sLORETA”, “LCMV”
plot_method_delays(
df,
["dSPM", "MNE", "sLORETA", "LCMV"],
xlim=[0, 0.15],
ylim=[0, 250]
)
plt.show()
/Users/payamsadeghishabestari/ANT/venv/lib/python3.10/site-packages/seaborn/axisgrid.py:123: UserWarning: Tight layout not applied. tight_layout cannot make Axes height small enough to accommodate all Axes decorations.
self._figure.tight_layout(*args, **kwargs)
/Users/payamsadeghishabestari/ANT/venv/lib/python3.10/site-packages/seaborn/axisgrid.py:123: UserWarning: Tight layout not applied. tight_layout cannot make Axes height small enough to accommodate all Axes decorations.
self._figure.tight_layout(*args, **kwargs)
/Users/payamsadeghishabestari/ANT/venv/lib/python3.10/site-packages/seaborn/axisgrid.py:123: UserWarning: Tight layout not applied. tight_layout cannot make Axes height small enough to accommodate all Axes decorations.
self._figure.tight_layout(*args, **kwargs)
/Users/payamsadeghishabestari/ANT/venv/lib/python3.10/site-packages/seaborn/axisgrid.py:123: UserWarning: Tight layout not applied. tight_layout cannot make Axes height small enough to accommodate all Axes decorations.
self._figure.tight_layout(*args, **kwargs)
Next, visualize delays for “eLORETA”
plot_method_delays(
df,
["eLORETA"],
xlim=[0.7, 1.1],
ylim=[0, 40]
)
plt.show()

Total running time of the script: (0 minutes 0.556 seconds)